Search
About Phaeodactylum tricornutum CCAP 1055/1
Phaeodactylum tricornutum is a diatom, the only species in the genus Phaeodactylum. Unlike other diatoms, P. tricornutum can exist in different morphotypes and changes in cell shape can be stimulated by environmental conditions. This feature can be used to explore the molecular basis of cell shape control and morphogenesis. Unlike most diatoms, P. tricornutum can grow in the absence of silicon and can survive without making silicified frustules. This provides opportunities for experimental exploration of silicon-based nanofabrication in diatoms.
Picture credit: Creative Commons Attribution 2.5 via Wikimedia Commons (Image source) Taxonomy ID 556484
(Text from Wikipedia.)
More information General information about this species can be found in Wikipedia
Taxonomy ID 556484
Data source Ensembl Protists
Comparative genomics
What can I find? Homologues, gene trees, and whole genome alignments across multiple species.
 More about comparative analyses
More about comparative analyses
 Phylogenetic overview of gene families
Phylogenetic overview of gene families
 Download alignments (EMF)
Download alignments (EMF)
Variation
What can I find? Short sequence variants.
 More about variation in Ensembl Protists
More about variation in Ensembl Protists
Other Data
P. tricornutum ESTs were sequenced from 16 different cDNA libraries. These libraries were constructed from cells cultured under different conditions in order to investigate the molecular basis for responses to various nutrient and stress conditions and changes in cell morphotype. These ESTs were aligned to the genome using exonerate.



 Display your data in Ensembl Protists
Display your data in Ensembl Protists
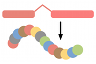
 Update your old Ensembl IDs
Update your old Ensembl IDs

![Follow us on Twitter! [twitter logo]](/i/twitter.png)
