Assembly mapping
Ensembl Genomes uses the A2Amapper, described below, to map between genome assemblies in different releases of Ensembl Genomes. The results of the mapping can be used to map data (in a variety of formats) to the current assembly, using the Assembly converter in the Tools section of the Ensembl website or via the Ensembl Core API.
Istrail S, Sutton GG, Florea L, Halpern AL, Mobarry CM, Lippert R et al. (2004) Whole-genome shotgun assembly and comparison of human genome assemblies. Proc Natl Acad Sci U S A 101 (7):1916-21. DOI: 10.1073/pnas.0307971100 PMID: 14769938
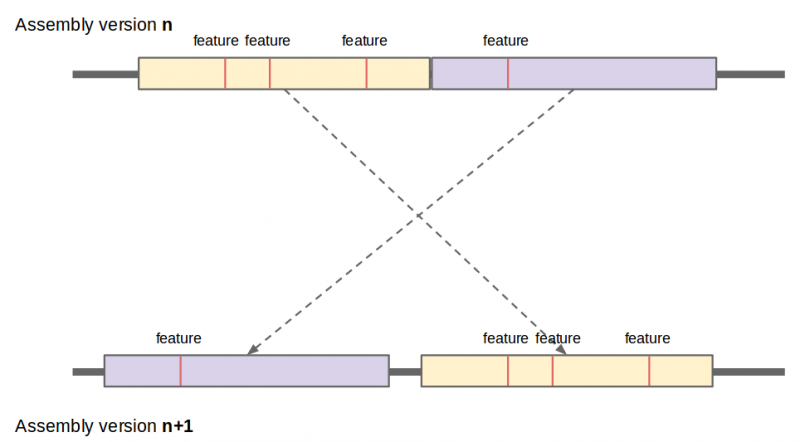
We have developed a suite of tools, A2Amapper, for constructing a one-to-one correspondence between pairs of assemblies. Like other whole-genome comparison methods, A2Amapper is based on the identification of seed alignments, in this case unique exact matches, followed by a more aggressive local alignment phase between seeds within nonoverlapping chains of seeds. Cutoffs were carefully tuned to balance sensitivity (finding all correlations), specificity (finding only the true ones), and computational requirements. A2Amapper produces a set of one-to-one matches that are alignments of nearly identical pairs of segments imputed to be analogous up to polymorphisms.
These large one-to-one matches are used to project position based features, such as variants between genome assembly versions.



![Follow us on Twitter! [twitter logo]](/i/twitter.png)
